Using Flow Cytometry to Immunophenotype Dendritic Cells: Master Regulators of the Immune System
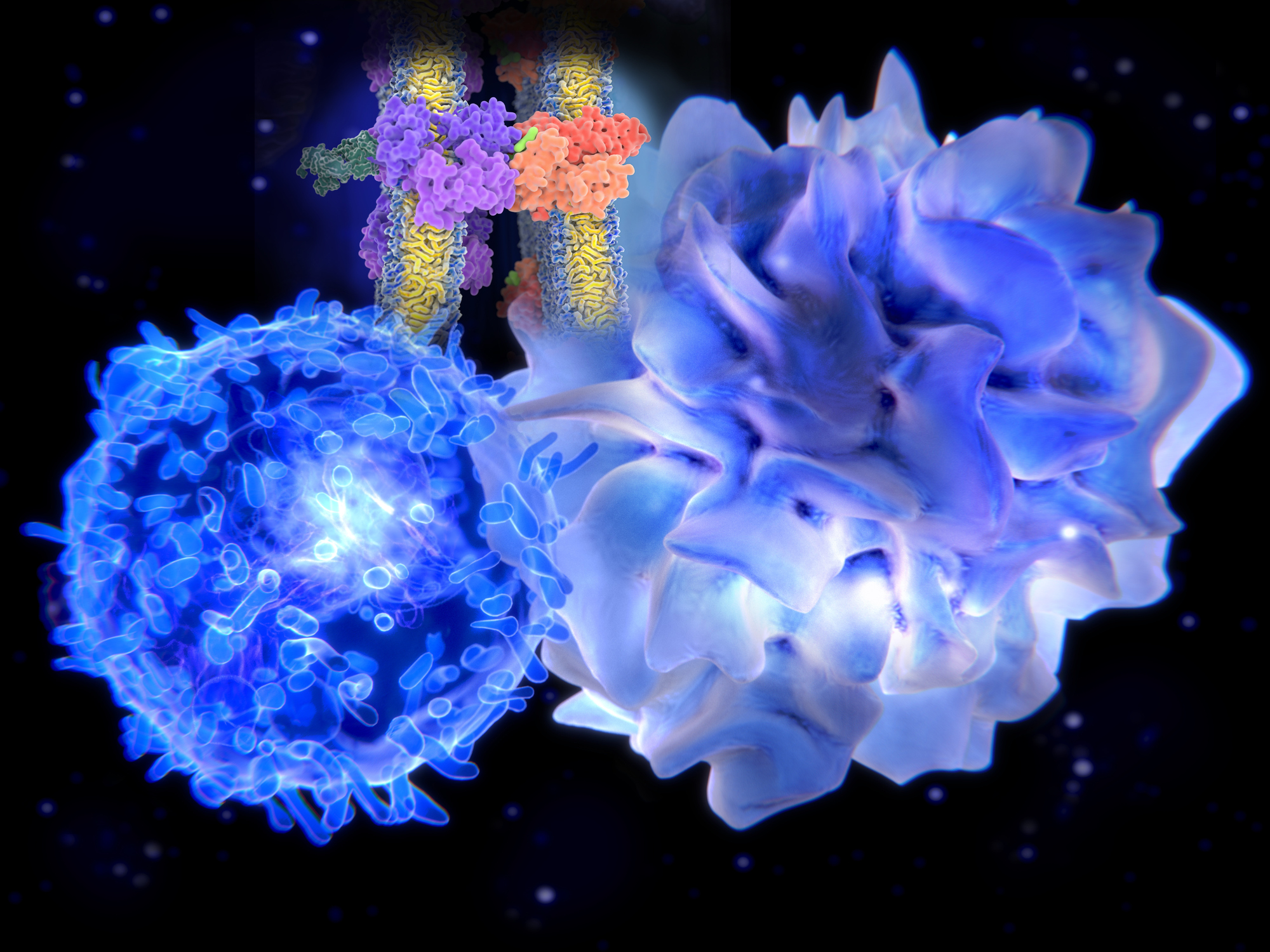
Dendritic cells (DC) are specialized antigen-presenting cells and key mediators of immunity and tolerance. DC bridge the innate and adaptive immune systems. As innate immune cells, DC recognize and respond to pathogen associated molecular Patterns (PAMPS) and damage/ danger associated molecular patterns (DAMPS) through toll-like receptors (TLR). The role of DC in adaptive immunity is to process proteins and present antigen in the MCH complex to prime naïve T cells (Figure 1). In addition, DC produce tolerogenic cytokines that induce the differentiation of Regulatory T cells. Subsets of DC express different markers, and are specialized in their response to pathogens. The unique phenotypes of DCs make them of considerable interest as potential therapeutic targets.

Figure 1 – The immune system consists of innate and adaptive immunity. Dendritic cells (DCs) bridge both innate and adaptive arms of the immune system. In an innate immune role, upon stimulation with PAMPs, DCs secrete cytokines that activate natural killer (NK) cells and also help to differentiate CD4+ and CD8+ T cells. In an adaptive immune role, upon encounter and internalization of antigen, DC load antigen-derived peptides onto Major Histocompatibility Complex (MHC) molecules. Peptides loaded onto MHC II (HLA-DR) molecules are recognized by CD4 T cells. Peptides loaded onto MHC I (HLA-ABC) molecules (a process called cross-presentation) are recognized by antigen-specific CD8 cells, driving their proliferation/activation/cytotoxicity. Figure from (Embgenbroich & Burgdorf, 2018).
DC are derived from the bone marrow, are represent about 0.5-1.5% of the peripheral blood mononuclear cells (PBMC). Although few in number, there are three specialized subsets of DC: 2 types of conventional, or classical, myeloid DC (cDC1 and cDC2), and plasmacytoid DC (pDC) (Breton et al., 2016). These specialized subsets can be defined by transcription factors, especially the interferon regulatory factors, IRF8 and IFR4. For example pDC express both IFR8 and IFR4, while cDC1 express IRF8 only and cDC2 express IFR4 only (Collin & Bigley, 2018).
All three DC subsets are derived from the bone marrow, and originate from a common Granuloycte, Monocyte and DC progenitor (GMDP) that develops into a monocyte–DC progenitor (MDP). MDPs in turn give rise to monocytes, and to a common DC precursor (CDP). CDPs can no longer give rise to monocytes, but give rise to all three subsets of DC. CDP are restricted to the bone marrow and give rise to pre-cDC and pDC. Pre-cDC cells migrate from the bone marrow to the blood or periphery to differentiate into cDC1 and cDC2 in lymphoid tissues (Figure 2). Recent studies have shown that pre-cDC are heterogeneous and are pre-committed to become either cDC1 or cDC2 based on CD172 (SIRP-α) expression (Breton et al., 2016).

Figure 2 – Dendritic Cell hematopoiesis. DC originate in the bone marrow (BM). Granulocytes, Monocyte, Dendritic Cell Progenitors (GMDP) develop into a monocyte and DC progenitor (MDP). MDPs give rise to monocytes and a common DC progenitor (CDP). CDP’s give rise to pDC and to circulating a cDC precursor (pre-cDC). Pre-cDC’s migrate from the bone marrow to the periphery, where they become cDC1 (BDCA-3) and cDC2 (BDCA-1). From (Breton, Lee, Liu, & Nussenzweig, 2015).
Briefly, some characteristics and functional attributes of the three DC populations are:
cDC1 cells (CD141+/BDCA-3) are present at 1/10 of the number of cDC2 cells in blood and tissue (lymph node, tonsil, spleen and bone marrow). cDC1 express some but not all myeloid markers (they express CD33, but lower levels of CD11c and little or no CD11b), and CLEC9A, a receptor involved in sensing necrotic cells, and the cross-presentation of antigen to activate CD8 cells and the TH1 response. cDC1 produce Type III interferon to induce and prime CD8 cells to recognize tumors and intra-cellular pathogens. Important cDC1 receptors include TLR3, TLR9 and TLR10.
cDC2 (CD1c+/BDCA-1) are the major DC population in blood and lymphoid organs. They express myeloid markers (CD11c, CD33, CD11b, CD13), and are primarily localized in the marginal zone of the lymph node and confer immunity against parasites, allergens, extra-cellular bacteria and fungi (Basit, Mathan, Sancho, & De Vries, 2018; Collin & Bigley, 2018). These cells excel at CD4 T cell priming and promote TH-17 and TH-2 immune responses. They are also capable of cross-presentation to CD8 cells. Important cDC2 receptors include TLR4, TLR6, TLR2/6, TLR8 and TLR9 (Breton et al., 2016; Collin & Bigley, 2018).
pDC (CD303+/BDCA-2) do not express myeloid markers, and retain the early progenitor markers CD123 (IL3-R) and CD45RA. pDC are localized to the extra-follicular, T cell rich areas of the lymph node, and are specialized to produce to Type 1 and Type II Interferons in response to viral infections. TLR7 and TLR9 are the key endosomal pattern recognition receptors for pDC (Collin & Bigley, 2018).
Flow Cytometry is the most commonly used method to identify DC in blood and lymphoid tissues. Flow cytometry can separate the three DC subsets using BDCA markers: Myeloid cDC1 express BDCA-3 (CD141), cDC2 express BDCA-1 (CD1c), and pDC express BDCA-2 (CD303) (Breton et al., 2015; Collin & Bigley, 2018; Collin, Mcgovern, & Haniffa, 2013). An example of a flow cytometry panel for immunophenotyping DC (and monocytes) is shown in Table 1, and a gating strategy in shown Figure 3.


Figure 3: Flow cytometry gating strategy for three DC subsets. HLA-DR+/Lineage negative (CD3/CD19/CD20/CD56) cells are gated and displayed by CD14 vs CD16, where different monocyte populations are identified (classical/non-classical and intermediate). From this plot, CD14-/CD16lo/CD16- cells are gated and displayed as HLA-DR by CD11c. HLA-DR+ /CD11c lo and CD11c+ cells are myeloid DC and can be further gated by BCDA-1 and BDCA-3 to identify cDC2 and cDC1 respectively. Finally, HLA-DR+/CD11c- cells are gated and displayed by HLA-DR by BDCA-2 (CD303), to identify plasmacytoid DC (pDC).
DC may be few in number, but they are key mediators of the immune system. Their biology continues to be an area of active investigation and they are the focus of drug development programs as potential therapeutic targets.
FCSL recognizes the evolving biology and ongoing research involving DC, and are privileged to support a number of drug development programs targeting DC. We are a contract flow cytometry lab and routinely run up to 10-color flow cytometry panels supporting basic research, non-clinical and clinical studies. We are experts in flow cytometry panel design, including fit-for-purpose flow cytometry panels, Immunophenotyping, Receptor Occupancy Assays (ROA), and the inclusion of proprietary markers into established flow cytometry panels. If your research involves DC, contact us to find out how flow cytometry can support your research.
References
Basit, F., Mathan, T., Sancho, D., & De Vries, J. M. (2018). Human dendritic cell subsets undergo distinct metabolic reprogramming for immune response. Frontiers in Immunology, 9(NOV), 1–17. https://doi.org/10.3389/fimmu.2018.02489
Breton, G., Lee, J., Liu, K., & Nussenzweig, M. C. (2015). Defining human dendritic cell progenitors by multiparametric flow cytometry. Nature Protocols, 10(9), 1407–1422. https://doi.org/10.1038/nprot.2015.092
Breton, G., Zheng, S., Valieris, R., da Silva, I. T., Satija, R., & Nussenzweig, M. C. (2016). Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. Journal of Experimental Medicine, 213(13), 2861–2870. https://doi.org/10.1084/jem.20161135
Collin, M., & Bigley, V. (2018). Human dendritic cell subsets: an update. Immunology, 154(1), 3–20. https://doi.org/10.1111/imm.12888
Collin, M., Mcgovern, N., & Haniffa, M. (2013). Human dendritic cell subsets. Immunology, 140(1), 22–30. https://doi.org/10.1111/imm.12117
Embgenbroich, M., & Burgdorf, S. (2018). Current concepts of antigen cross-presentation. Frontiers in Immunology, 9(JUL). https://doi.org/10.3389/fimmu.2018.01643

